Free.Servers
SwiftLib
SwiftLib degenerate codon library optimizer
Recent.Publications
Using anchoring motifs for the computational design of protein-protein interactions.
Jacobs TM, Kuhlman B. Biochem Soc Trans. 2013 Oct 1;41(5):1141-5.
Adding diverse noncanonical backbones to rosetta: enabling peptidomimetic design.
Drew K, Renfrew PD, Craven TW, Butterfoss GL, Chou FC, Lyskov S, Bullock BN, Watkins A, Labonte JW, Pacella M, Kilambi KP, Leaver-Fay A, Kuhlman B, Gray JJ, Bradley P, Kirshenbaum K, Arora PS, Das R, Bonneau R. PLoS One. 2013 Jul 15;8(7):e67051
Rosetta.design3.5
computational protein design softwarewelcome.
Rosetta design can be used to identify sequences compatible with a given protein backbone. Some of Rosetta design's successes include the design of a novel protein fold, redesign of an existing protein for greater stability, increased binding affinity between two proteins, and the design of novel enzymes.
If you would like to use Rosetta.design, please register for an account. If you already have an account, you can login below.
login.
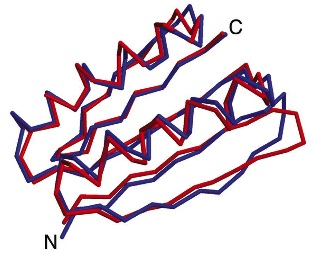
crystal structure of Top7
Comparison of the computationally designed model (blue) to the solved x-ray structure (red) of Top7 reveals a backbone RMSD of only 1.17A.